 According to the evolutionary paradigm, complex genetic information in the form of genes and regulatory DNA can randomly evolve through mutations and selection. But this erroneous idea becomes more untenable with every new discovery in the field of genomics.
According to the evolutionary paradigm, complex genetic information in the form of genes and regulatory DNA can randomly evolve through mutations and selection. But this erroneous idea becomes more untenable with every new discovery in the field of genomics.
For example, research on meiosis, a type of cell division that produces genetic variation when sperm and egg cells form, is creating major roadblocks for conventional evolutionary theory. Genetic recombination, a key event during meiosis, is proving to be especially problematic for the mutation-selection myth. The precise regulation of this process, once thought to be largely random, involves an amazing amount of engineering.
Meiosis, an Introduction
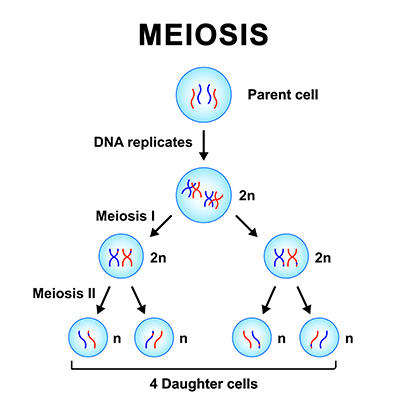
For sexually reproducing organisms, the cellular division process of meiosis plays a vital role in generating genetic diversity. This is not to be confused with mitosis, which involves replication of body (somatic) cells during normal growth and development.1
Meiosis essentially has two missions in facilitating gametogenesis (sperm and egg production). The first is genome reduction, in which the normal cellular chromosome content—diploid (2n) in most cases, with two complete sets of chromosomes—is reduced to a haploid (1n) with one set of chromosomes. When a haploid egg and a haploid sperm combine in fertilization, the diploid or 2n normal state of the genome is reconstituted. This is why people have two sets of chromosomes, one from the father (paternal genome) and one from the mother (maternal genome).
This article will focus on the second goal of meiosis: creating genetic variation in offspring. Creating genetic diversity or variation is a key feature in maintaining animal and human health. The opposite of this concept is demonstrated by the higher rate of birth defects that are commonly observed when genetic diversity is lost due to inbreeding.

The way that meiosis generates this genetic variation is actually quite an ingenious and intriguing process since it involves two separate types of randomization/shuffling.
The first phase of randomization is a form of literally shuffling the genome like a deck of cards on a massive scale. Called recombination, this absolutely amazing event could be completely fraught with extreme hazard if governed by anything short of the most precise engineering. It first involves the pairing of homologous chromosomes. In other words, chromosome 1 inherited from the father pairs with chromosome 1 inherited from the mother, and so on.
In the human genome, 22 regular chromosomes (autosomes) pair up in a perfect fashion. Once this is done, the homologous chromosomes literally begin “crossing over” each other in the recombination process. Highly efficient cell machinery slices, dices, and reconnects fragments of DNA back and forth between the paired chromosomes in what is thought to be a random but controlled manner.
The end result is that the maternal and paternal chromosomes become new chimeric or recombined chromosomes, having exchanged numerous segments with each other. Although the genes and various other DNA features have been exchanged/recombined, the linear order and integrity of these components are preserved throughout the whole process. This is one of recombination’s remarkable but necessary attributes that maintain genome stability and function.
The second level of genetic variation immediately follows recombination. In this phase, the chromosomes are pulled apart to form two separate sets in a process called independent assortment. This means a newly recombined chromosome has a 50% chance of ending up in one set or the other. It offers an additional level of randomization, helping to increase and maximize the available genetic diversity that the process can generate. The randomization from shuffling the genetic deck of cards twice is the reason most siblings look different from each other even though they have the same parents.

Meiosis is basically a one-way continuous process. It’s not part of a stop-and-go cell cycle system like mitosis. In addition, meiosis only takes place in the reproductive organs of plants and animals, not in the other tissues. Its entire goal is to create new genome combinations and promote the genetic diversity needed to safeguard against the effects of inbreeding.
Recombination or crossover frequency is typically determined by DNA sequencing the haploid genomes of gametes. For example, a recent study in water buffalo involved the DNA sequencing of 78 sperm cells from a single buffalo. The researchers were then able to identify 1,956 crossovers with an average of 25.1 per sperm cell, which is similar to human studies.2
Recombination, a Highly Engineered System
One of the initial molecular findings about recombination was that it is not random but instead occurs in a highly regulated manner. First, researchers discovered that recombination events tend to be unevenly distributed around the genome and frequently occur in small, specified genomic regions termed recombination hotspots.3 Interestingly, these regions are characterized by several DNA signatures.

For example, these sections have a higher-than-average amount of guanine (G) and cytosine (C) DNA bases compared to the amount of adenine (A) and tyrosine (T)—known as GC content. In addition to an elevated GC content, these areas are commonly associated with a DNA feature called transposable elements (TEs), which I’ve discussed in previous articles.4,5 There is typically a negative association between recombination and TEs because the more TEs that are present in a chromosomal region, the less recombination tends to occur.
Finally, a specialized DNA binding protein called PRDM9 has been found to localize almost all meiotic recombination sites in humans and mice, but most PRDM9-bound DNA segments themselves do not become recombination hotspots. 6 However, the oddity of many different creatures either having or not having a functional PRDM9 protein is a complete evolutionary enigma.7
For example, while mice and humans have PRDM9, platypuses and dogs do not. Other examples are found among types of ray-finned fish, amphibians, reptiles, and birds. In other words, PRDM9 is an important protein for regulating recombination in some creatures, but in others it’s not utilized. Its presence or absence in the scheme of life negates evolution and supports the specificity of creature kinds by divine creation.
Gene Control Regions Are Protected, Negating Evolution
Recombination hotspots among mammals are poorly conserved. This means their locations in the genome and DNA sequence structure are unique to creature kinds and don’t support the evolutionary theory of one fundamental creature kind morphing into another.8 The common house mouse (Mus musculus) is one of the best mammalian models for studying recombination. Recombination hotspots have been mapped all over the mouse genome.8,9
One key research finding is that genetic recombination is directed away from sensitive parts of the genome that contain genetic control elements and features.10 These parts of the genome carefully regulate how genes are turned off and on and how they function in precisely regulated networks.
In creatures that lack the PRDM9 protein, though, recombination does occur in regulatory regions of genes. Researchers think this is facilitated by the fact that such regions tend to naturally contain open and active DNA. Furthermore, the chromosomes in many bird genomes are very small and compact, lacking large amounts of transposable elements as well as the PRDM9 protein.7 And while recombination occurs consistently in regulatory regions in genomes that lack PRDM9, it does not occur within the gene bodies themselves.
Epigenetics and Transposable Elements
Epigenetics is one of the major factors that regulate and control recombination.11 I discussed the basic systems of epigenetics in previous articles: DNA methylation, histone modifications, and small RNAs.12,13 Epigenetic modifications strongly regulate crossover positioning by altering the accessibility of DNA to the recombination machinery. The process of homologous chromosome pairing during meiosis prior to recombination also influences the positioning of crossovers.
Transposable elements (TEs) also help govern recombination.4 As briefly noted above, one of the most striking patterns of genome structure is the strong, typically negative association between TEs and recombination rates. In other words, the denser a region of a genome is in TEs, the less recombination occurs. While this appears to be a common feature of eukaryotic genomes, the mechanisms driving the strong correlation between TEs and recombination are poorly understood.

In addition, and more rarely, this association can be totally reversed, a phenomenon typically associated with the types of TEs involved. For example, there is contrasting evidence in humans depending on TE type. One type, called L1 LINEs, is negatively correlated with recombination rates, while another type of TE, called Alu elements, tends to predominate in gene-rich, high-recombination regions.
Recombination and Environmental Cues
Interestingly, recombination is responsive to both intrinsic and environmental factors. Intrinsic factors include the sex and age of the organism. In many plants and animals, including mice and humans, females tend to have higher rates of recombination than males.14 As organisms grow older, recombination rates tend to decline. In humans, mice, fruit flies (Drosophila), and nematodes (Caenorhabditis elegans), the effects of sex and age intersect with recombination rates decreasing with maternal age.

In regard to environmental responses, the most well-studied paradigm is recombination effects due to changes in temperature. 14 In a wide variety of organisms—e.g., Arabidopsis thaliana (a small weedy plant), Hordeum vulgare (barley), nematodes, grasshoppers (Melanoplus femurrubrum), and Hyacinthus orientalis (an ornamental garden plant)—an increase in recombination frequency with increasing temperature is well documented. However, the temperature response has built-in thresholds in which meiosis is shut down when the temperature gets too cold or too hot.
Conclusion
Evolutionists have speculated for years that mutation combined with homologous recombination is one of the key mechanisms of evolutionary change. They claim it operates as some sort of mystical tinkering mechanism that miraculously generates novel genes that somehow become fully and precisely integrated into the genome’s functional networks. The emerging concept that homologous recombination is a controlled feature of the genome limited to specific hotspots contradicts the idea of random evolutionary processes being able to produce new genes and regulatory DNA sequences.
There is even an anti-evolutionary angle to this when it comes to the human-ape DNA nonsimilarity issue. When researchers compared recombination in chimpanzees to that in humans, they found that “chimpanzee recombination is dominated by hotspots, which show no overlap with humans even though rates are similarly elevated around CpG islands and decreased within genes.”15 This is to be expected because the chimpanzee and human genomes are turning out to be much more different than scientists originally predicted—negating evolution.16, 17
Overwhelming scientific data show that the entire process of gamete formation, including recombination, is a highly engineered and precisely governed system that speaks directly to its all-wise and all-powerful Creator, the Lord Jesus Christ.
References
- I have reviewed both mitosis and meiosis in depth in a previous book. See Tomkins, J. P. 2018. Cell Division and DNA Replication: How Life is Engineered to Perpetuate. In The Design and Complexity of the Cell. Dallas, TX: Institute for Creation Research, 57–67.
- Wang, X. et al. 2023. Chromosome-Level Genome and Recombination Map of the Male Buffalo. GigaScience. 12: 1–10.
- Clément, Y. and P. F. Arndt. 2013. Meiotic recombination strongly influences GC-content evolution in short regions in the mouse genome. Molecular Biology and Evolution. 30 (12): 2612–2618.
- Kent, T. V., J. Uzunovi?, and S. I. Wright. 2017. Coevolution between transposable elements and recombination. Philosophical Transactions of the Royal Society B: Biological Sciences. 372 (1736): 20160458.
- Tomkins, J. P. 2023. Transposable Elements: Genomic Parasites or Engineered Design? Acts & Facts. 52 (9): 14–18.
- Altemose, N. et al. 2017. A map of human PRDM9 binding provides evidence for novel behaviors of PRDM9 and other zinc-finger proteins in meiosis. Elife. 6: e28383.
- Bascón-Cardozo, K. et al. 2024. Fine-Scale Map Reveals Highly Variable Recombination Rates Associated with Genomic Features in the Eurasian Blackcap. Genome Biology and Evolution. 16 (1).
- Neale, M. J. and S. Keeney. 2006. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 442 (7099): 153–158.
- Smagulova, F. et al. 2011. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 472 (7343): 375–378.
- Brick, K. et al. 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature. 485 (7400): 642–645.
- Chowdary, K. V. S. K. A., R. Saini, and A. K. Singh. 2023. Epigenetic Regulation During Meiosis and Crossover. Physiology and Molecular Biology of Plants. 29 (12): 1945–1958.
- Tomkins, J. P. 2023. Epigenetic Mechanisms: Adaptive Master Regulators of the Genome. Acts & Facts. 52 (4): 14–17.
- Tomkins, J. P. 2024. Small Heritable RNAs Pack a Big Adaptive Punch. Acts & Facts. 53 (1): 12–15.
- Modliszewski, J. L. and G. P. Copenhaver. 2017. Meiotic Recombination Gets Stressed Out: CO Frequency Is Plastic Under Pressure. Current Opinion in Plant Biology. 36: 95–102.
- Auton, A. et al. 2012. A Fine-Scale Chimpanzee Genetic Map from Population Sequencing. Science. 336 (6078): 193–198.
- Tomkins, J. P. 2023. Human-Chimp DNA Similarity Research Refutes Evolution. Acts & Facts. 52 (10): 8–11.
- Tomkins, J. P. 2020. Human Chromosome 2 Fusion Never Happened. Acts & Facts. 49 (5): 16–19.
* Dr. Tomkins is a research scientist at the Institute for Creation Research and earned his Ph.D. in genetics from Clemson University.



