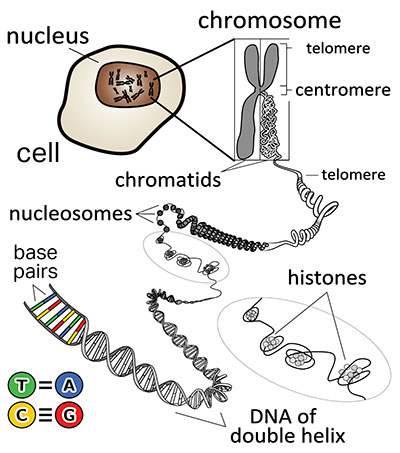
In eukaryotes, which are organisms with nucleated cells, the vast majority of hereditary and coded information is stored, copied, and replicated in the chromosomes within the nucleus.1 In human beings, the genetic (chromosomal) material within a single cell is about six feet in length. This needs to fit within an incredibly small space, so our Creator designed an elaborate packaging system to organize and compact the DNA of a chromosome at multiple levels.
In the design solution for this challenging spatial problem, the genome must be organized and compacted in such a way that cellular machinery in the nucleus can properly access the DNA. This allows the implementation of numerous gene expression programs that need to be executed at the right time and in the right tissue and cell type. This arrangement couldn’t have come about through random, hit-or-miss natural processes. It must have been engineered from the beginning.
Exploring the Function and Structure of the Genome
Various microscopy techniques have documented an array of features that provides insights into how the genome is organized in a cell’s nucleus. One technique called chromosome painting involves using specific fluorescent probes that light up a single chromosome.2 After highlighting individual chromosomes, a color can be assigned to each one. This generates a composite image of the nucleus showing a chromosome’s specific territory. Figure 1 depicts the territory for each human chromosome in a fibroblast cell.3

Other technologies zoom in and provide even more detailed information about the structure of the genome beyond the level that microscopy reveals. Most of these approaches are based on a technique called crosslinking that chemically fixes (binds together) areas of chromosomes that interact with each other.4,5 These crosslinked, interacting sections of different chromosomes can be isolated and their DNA sequenced.
This DNA sequence can then be used to determine the locations or addresses on each of the interacting chromosomes that were captured together during the crosslinking phase. The various types of interreacting genes or regulatory switches can be determined from the DNA sequence. This technology is called chromosome conformation capture and can involve a wide variety of technical variations.
Genome Architectural Design
Before getting into some of the principal functional features of the 3-D genome, another important point to discuss is the various levels of organization in the way the DNA is packaged and structured.3 Eukaryotic genomes are organized via several common architectural design features. The basic organizational elements of the genome are the DNA fibers, loops, domains, and compartments that chromosomes form.
The packaged and organized DNA of a chromosome is called chromatin. A chromatin fiber contains a stretch of 146 base pairs of DNA wrapped around a cluster of eight proteins called histones (Figure 2). Each one of these so-called “beads on string” are termed nucleosomes. This packaged form of DNA can be highly condensed or folded into higher-order features, such as chromosomal territories or domains, as noted above.
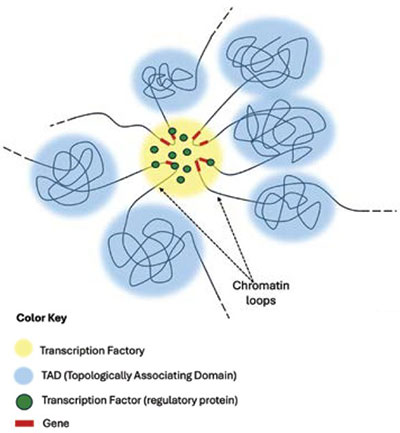
The Specific Locality of Genes and Transcription Factories
As mentioned earlier, chromosomes occupy specific regions within the nucleus. These are termed chromosome territories, and they can be further subdivided into specified chromosomal compartments referred to as topologically associating domains (TADs). Here the chromosomes can form loops that interact with loops from other chromosomal regions (Figure 3). These loops then bring together a group of genes that are used in a common cellular process in what is called a transcription factory (Figure 3).6,7
In light of all this, it’s important to understand that active genes and regulatory sequences are not randomly dispersed within the nucleus. It is now widely accepted that the location of co-regulated genes on different chromosomes is not accidental but highly specified since they occupy the same functional locations within the nucleus. This well-documented observation undergirds the concept of nuclear territories, in which a section on a particular chromosome exhibits repeated spatial connections with other chromosomal sections in response to a certain cell type or a specific functional process, such as those related to cellular and organism adaptive responses.
In a previous article, I explained how alternative splicing generates incredible diversity in gene products from a single gene.8 A single gene can produce over a hundred different messenger RNA variants (transcripts). Another interesting feature of alternative splicing is that it can also occur between messenger RNAs produced between genes located on completely different chromosomes.
The transcription factory regions and the machinery within them appear to remain relatively stationary while the DNA is moved into place and reeled through like film through a projector. For all of this to occur, genes in different regions of the genome are dynamically positioned within close physical proximity to each other and are transcribed in highly complex gene factory zones, as noted above.9–13 This is why gene activity (transcription) is not widely dispersed throughout the nucleus but is gathered in the transcription factories where approximately 10 genes at a time can be engaged and co-regulated.
The 4-D Nucleome
Not only is the genome arranged in highly ordered and specified configurations in three dimensions within the cell nucleus, but the dimension of time can be added to the mix as well. In this respect, researchers are trying to integrate the complexity of the three-dimensional aspect of the genome (spatial) with its activity in time (temporal) in various tissues and in response to external stimuli. This involves simultaneously studying thousands of genes and their physical positions in the genome using a variety of genomic technologies in which the data must be integrated and analyzed with elaborate statistical models.
In a 2015 study, researchers performed part of their analyses using the chromosome confirmation capture technique, which I mentioned in a previous article.14 They combined these results with additional information from 3-D microscopic imaging and millions of messenger RNA sequences captured across the genome to develop an integrated picture of activity in the 3-D genome in the nucleus (nucleome).
As if three dimensions were not enough, the researchers repeated these experiments across multiple time points in response to the body’s day/night timekeeping system, called a circadian clock, thus adding the fourth dimension of time to the study. Amazingly, they discovered thousands of genes across the 3-D genome dynamically and precisely regulated by the body’s internal clock, a stunning orchestration of complex genetic activity. The researchers wrote,
Genomic movements in 3D space provide a geometric picture of gene regulation in the context of circadian clocks, one that may give insight into the mechanisms regulating biological time.14
Not only are thousands of genes coordinately regulated together in 3-D space in a precise manner according to cell type and relevant physiological processes, but they also function within the context of time, the fourth dimension, in a wondrously precise genetic dance.
Studies like this actually led to the establishment of a federally funded 4-D nucleome project at the U.S. National Institutes of Health (NIH).15 The agenda for this was initially stated in a high-profile scientific paper published in 2017, in which the authors wrote:
To determine how the genome operates, we need to understand not only the linear encoding of information along chromosomes, but also its three-dimensional organization and its dynamics across time, that is, the ‘4D nucleome.’16
In a 2024 paper, a group of largely European scientists describe how an international 4-D nucleome project funded in part by the European Union was also started in 2015. It works alongside the NIH-funded project in the U.S. The authors wrote, “It is essential to visualize the genome in space and time.”17
Adaptation and 3-D Genomic Response
So, what role does the 3-D genome play in the adaptation of living creatures? Amazingly, scientists have recently been able to decipher the dynamics of the 3-D structure of the genome in a diversity of creatures that can live in a variety of environments that provide extreme adaptive challenges.
Oysters are ecologically and economically important bivalve mollusks that inhabit the wide part of a river where it interfaces with the ocean (estuarine) and coastal regions where the ocean meets the land between high and low tides (intertidal zones). These environments experience sharp fluctuations in temperature and salinity because of the tide cycle.
In addition, oysters are sessile creatures, incapable of active movement since the mature oyster will cement itself to rocks. Because of these factors, oysters and other intertidal or shallow sea organisms have been designed with high levels of genetic diversity and built-in plasticity to cope with rapid environmental changes. Thus, they make excellent models for adaptation and genomic responses.
In a recent study, researchers analyzed the 3-D genome architectural response of oysters that were transferred from the northern coast of China to the southern coast of China, two locations with radically different environments.18 Amazingly, the translocated oyster immediately underwent significant changes in its 3-D genome structure that created new TADs and areas of active gene function in response to environmental conditions, especially water temperature.
Not only were new, active regions of DNA configured, but the researchers found that several chromosomes were highly specialized to respond to environmental change.18 These chromosomes had designated 3-D response regions that exhibited a large amount of transcriptional adaptive plasticity. And many of the genes involved in the adaptive responses were related to metabolism and muscle movement that not only enable the oysters to rapidly reconfigure their metabolism but also alter their ability to close and open their shells. Another standout feature of this research is that many regions of the oyster genome that contained areas lacking protein-coding genes were critical to the formation and regulatory activity of the 3-D genome. This provides more evidence to debunk the myth of junk DNA.19

While the oceans offer challenging environments for life, so does land. In another recent study, researchers analyzed the 3-D genome in chickens that have the ability to adapt to both warm and humid tropical environments and cold and frigid northern locations.20 Because of their short growth and reproductive cycles and their wide geographic distribution, chickens are ideal model creatures for studying adaptive genome modifications.
The researchers chose to examine liver cells in two different breeds of chickens—one adapted to the tropics and one adapted to frigid conditions.20 The results showed that in response to its environment, each chicken had developed unique 3-D genome configurations with different TADs. These environment-specific TADs allowed for precise sets of genes to be expressed in transcription factories specifically tailored to each environment. The variability in gene pathways was connected to traits such as heat tolerance, cold adaptation, immune responses, and metabolic changes. In adaptation to cold and hypoxic conditions, gene pathway alterations were associated with smooth muscle contraction, vasoconstriction, and vasodilation.
Conclusion
Genes and regulatory elements are not randomly dispersed within the cell’s nucleus. They occupy precise locations with respect to the nuclear meshwork of structural fibers (lamina) and to each other. This important observation provides the fundamental basis of the well-accepted concept of highly specified and regulated nuclear territories.
These specialized chromosomal configurations exhibit reproducible connections within space and time according to cell type and even inputs from environmental conditions related to adaptation. As a result, gene transcription is not widely or randomly dispersed throughout the nucleus but is organized into specified nuclear sites called transcription factories that typically accommodate about 10 different genes concurrently transcribed as a group. And if this were not complex enough, this incredible activity is adaptably responsive and variable in time.
No random processes could result in such optimal function and intricate organization. This phenomenal complexity can only be attributed to the engineering of an all-wise and all-powerful Creator, the Lord Jesus Christ.
References
- Tomkins, J. P. 2018. The Design and Complexity of the Cell. Dallas, TX: Institute for Creation Research.
- Meaburn, K. J. and T. Misteli. 2007. Chromosome Territories. Nature. 445 (7126): 379–781.
- Fibroblasts produce the structural framework for tissues and organs.
- Jerkovic, I. and G. Cavalli. 2021. Understanding 3D Genome Organization by Multidisciplinary Methods. Nature Reviews. 22: 511–528.
- Perillo, B., A. Migliaccio, and G. Castoria. 2024. Chromatin Looping Links Gene Expression to the Assembly of Transcription Factories (Review). Molecular Medicine Reports. 29 (6): 97.
- Li, H. B. et al. 2013. Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies. PLoS Genetics. 9 (4): e1003436.
- Perillo, Migliaccio, and Castoria, Chromatin Looping.
- Tomkins, J. P. 2025. Gene Complexity Showcases Engineered Versatility. Acts & Facts 54 (1): 14–17.
- Van Bortle, K. and V. G. Corces. 2012. Nuclear Organization and Genome Function. Annual Review of Cell and Developmental Biology. 28: 163–187.
- Davidson, S., N. Macpherson, and J. A. Mitchell. 2013. Nuclear Organization of RNA Polymerase II Transcription. Biochemistry and Cell Biology. 91 (1): 22–30.
- Pan, Q. et al. 2008. Deep Surveying of Alternative Splicing Complexity in the Human Transcriptome by High-Throughput Sequencing. Nature Genetics. 40 (12): 1413–1415.
- Yustis, J. C., M. Devoucoux, and J. Cote. 2024. The Functional Relationship Between RNA Splicing and the Chromatin Landscape. Journal of Molecular Biology. 436 (16).
- Wang, E. T. et al. 2008. Alternative Isoform Regulation in Human Tissue Transcriptomes. Nature. 456 (7221): 470–476.
- Tomkins, J. P. Human Nucleome Reveals Amazing 4-D World. Creation Science Update. Posted on ICR.org July 27, 2015. See Chen, H. et al. 2015. Functional Organization of the Human 4D Nucleome. Proceedings of the National Academy of Science of the United States of America. 112 (26): 8002–8007.
- The 4D Nucleome Program. National Institutes of Health. Posted on 4dnucleome.org.
- Dekker, J. et al. 2017. The 4D Nucleome Project. Nature. 549 (7671): 219–226.
- Rey-Millet, M. et al. 2024. The Genome in Space and Time Comes of Age. Nucleus. 15 (1): article 2307665.
- Li, A. et al. 2023. Genome Architecture and Selective Signals Compensatorily Shape Plastic Response to a New Environment. The Innovation. 4 (4): 100464.
- Tomkins, J. P. Human Genome 20th Anniversary…Junk DNA Hits the Trash. Creation Science Update. Posted on ICR.org April 12, 2021.
- Shao, D. et al. 2022. Three-Dimensional Organization of Chicken Genome Provides Insights into Genetic Adaptation to Extreme Environments. Genes. 13 (12): 2317.