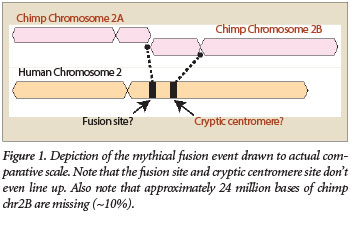
 One of the most common arguments evolutionists use to promote the theory that humans evolved from an apelike ancestor is the idea of a “chromosome 2 fusion.” This story proposes that in a common ancestor shared by humans and chimps, two small chromosomes somehow fused end to end to produce human chromosome 2 (Figure 1).1 This supposedly explains the difference in chromosome numbers between humans and great apes—humans have 46 chromosomes, while great apes such as chimps, gorillas, and orangutans have 48.
One of the most common arguments evolutionists use to promote the theory that humans evolved from an apelike ancestor is the idea of a “chromosome 2 fusion.” This story proposes that in a common ancestor shared by humans and chimps, two small chromosomes somehow fused end to end to produce human chromosome 2 (Figure 1).1 This supposedly explains the difference in chromosome numbers between humans and great apes—humans have 46 chromosomes, while great apes such as chimps, gorillas, and orangutans have 48.
To understand the concept associated with an end-to-end fusion model, it is important to know what the ends of chromosomes, telomeres, look like. Telomeres are made up of special chromosomal end sequences of the six-base DNA sequence TTAGGG that are repeated over and over again in perfect tandem. In fact, typical human telomeres are quite large, between 5,000 to 15,000 bases long.2
In 1991, scientists reported finding alleged evidence of a telomere-telomere fusion event on human chromosome 2, but it was not quite what they were expecting.3 First, the sequence was only about 800 bases long—not the 10,000 bases or more you would expect if two 5,000-base (or larger) telomeres fused together.
Second, the fusion-like sequence was very degenerate and only 70% similar to what one would expect of a pristine fusion sequence of the same size. Even if you assume an evolutionary timeline of up to six million years since the fusion event occurred, the data do not match up with known mutation rates or the variability found in human DNA.
A third major problem is the fusion site contains no type of sequence called satellite DNA (satDNA). In chromosome fusion events that occur in nature in living mammals—a very rare event—the DNA signature always involves satDNA producing a DNA signature that occurs as either satDNA-satDNA or satDNA-teloDNA sequence.4,5,6 Thus, the alleged fusion event should contain satDNA—a problem the fusion site discoverers openly acknowledged in their initial 1991 paper. When teloDNA-teloDNA fusions do occur in humans, they involve tissues and cell lines associated with cancerous tumors.7,8,9
A fourth major problem for the alleged fusion signature on chromosome 2 is that it occurs in a region of the genome that is full of genes. Telomeres do not contain genes, yet the fusion site is in the midst of a hotbed of genetic activity. The gene neighborhood surrounding the alleged fusion lacks overall synteny (similar gene order) to the chimp genome and does not support a fusion scenario in any way. This was first noticed in 2002 by secular researchers, although the chimp genome had not been well sequenced at that time.10,11 This author has recently verified that an overall lack of synteny supporting fusion still holds true for over 2.7 million bases surrounding the fusion site based on the most recent version of the chimpanzee genome compared to human (Tomkins, unpublished data).
Despite all of these serious difficulties, the greatest problem that evolutionists now have is the fact that the alleged fusion sequence is located in the middle of a functional gene.12 It is not a fossil remnant of a chromosomal accident at all but an important DNA regulatory feature called a promoter (genetic switch) inside a highly expressed gene.
More specifically, the purported fusion site is located inside a crucial RNA helicase gene called DDX11L2 that produces long non-coding RNAs. This gene is expressed in at least 255 different tissue and cell types in humans and is highly coregulated with many other important genes in the cell.12 So not only is the gene highly active throughout the human body, it is tightly networked with many other genes, including those that are involved in the development of blood cells.
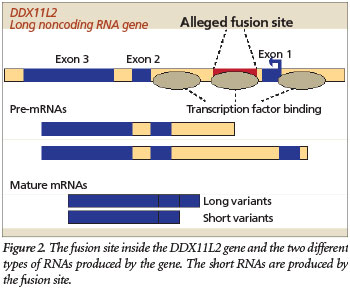 But the evolutionary fusion story gets worse. The fusion-like sequence itself has an important functional purpose based on recent data available at the UCSC Genome Browser (genome.ucsc.edu) genomic database. Specifically, the fusion site sequence binds to at least 11 different transcription factors, including RNA polymerase II, the key enzyme that transcribes genes. Transcription factors are specialized proteins that turn genes off and on. The fact that these proteins specifically bind to the alleged fusion site sequence indicates that it is a promoter located inside the gene (Figure 2). It is common for human genes to have these promoter regions located both in front of the main body of the gene and inside them.
But the evolutionary fusion story gets worse. The fusion-like sequence itself has an important functional purpose based on recent data available at the UCSC Genome Browser (genome.ucsc.edu) genomic database. Specifically, the fusion site sequence binds to at least 11 different transcription factors, including RNA polymerase II, the key enzyme that transcribes genes. Transcription factors are specialized proteins that turn genes off and on. The fact that these proteins specifically bind to the alleged fusion site sequence indicates that it is a promoter located inside the gene (Figure 2). It is common for human genes to have these promoter regions located both in front of the main body of the gene and inside them.
In the case of the fusion-like sequence, its designed function is as a second promoter inside the DDX11L2 gene. As mentioned above, RNA polymerase II also binds to the fusion sequence. Along with the binding of RNA polymerase is the fact that transcription has also been shown to start inside the fusion-like sequence, further validating it as a functional genetic element inside an important gene, not some random accident of fusion. This data can be found in the Functional Annotation of the Mammalian Genome (fantom.gsc.riken.jp/5) database and will be described in more detail in a future technical report by this author.
So, what is the ultimate designed purpose of this supposed fusion-like sequence? First, it is important to note that DNA is a double-stranded molecule and genes are located on both of its strands, running in opposite directions from each other. The two DNA strands are typically designated the plus and minus strands. The DDX11L2 gene and the fusion-like sequence inside it are actually encoded on the minus strand and oriented in the reverse complement of what is typically depicted as the fusion sequence. Second, the DDX11L2 gene actually encodes RNA transcripts of two different lengths—short variants (~1,700 bases long) and long variants (~2,200 bases long). In this respect, the fusion site itself appears to be the functional start site for the shorter transcript variants (Figure 2).
Another very interesting fact about this sequence is that it is not entirely unique to the purported fusion site. Portions of it contain significant levels of similarity to other areas throughout the human genome, including many internal regions that are not near telomeres.13,14 These internally located telomere-like sequences in the middle of chromosomes are quite common in the human genome and are believed to serve various functional purposes, including the regulation of gene expression.15,16
While evolutionists typically point to the presence of an alleged fusion site as the main evidence of fusion, they also claim that a cryptic centromere exists as well. However, the evidence for this is considered to be much weaker. The general idea is based on the fact that if a fusion of two smaller chromosomes actually occurred in the distant past, you would have two centromeres in the newly formed chimeric chromosome—one from each of the two parent chromosomes (Figure 1). Centromeres are specialized sections of DNA located inside chromosomes that help cells control the DNA during cell division. In the case of a fusion event, this would instantly create an unstable situation leading to the destruction of the cell. According to the evolutionary fusion model, one of the two centromeres in the newly fused chromosome was somehow “silenced” and then degenerated over time.
Centromeres are composed of a type of DNA sequence called alphoid DNA, a clearly recognizable DNA sequence pattern about 171 bases in length. Alphoid DNA is actually found all over the human genome, but it is important to understand that there are different types (variants) of alphoid DNA sequence, each with different functions in the genome.
Research has shown that the alphoid DNA alleged to be a cryptic centromere is not the same type as that found at actual functional human centromeres.17 The alphoid sequences at the supposed cryptic centromere site are quite diverse and form three separate subgroups.13 In fact, the overall patterns of the alphoid sequence sub-members are not even similar to the patterns found in functional human centromeres. In addition, the location of the alleged cryptic centromere site does not match where it should be if a fusion actually occurred (Figure 1).
Complicating matters further is the fact that alphoid sequences differ markedly between humans and apes.18,19 In fact, when the human alphoid DNA sequence at the supposed ancient centromere site is compared to the chimp genome, not a single similar positive match is obtained.13 Thus, not only is the so-called fossil centromere sequence dissimilar to functional human centromeres, it has no sequence correspondence to the chimp genome either.
In short, the overwhelming lack of evidence for a chromosome 2 fusion does not support the evolutionary story in any feasible way. Clearly, the so-called fusion sequence is an important functional feature called a promoter inside a highly expressed gene, not an accident of evolution.
References
- Yunis, J. J. and O. Prakash. 1982. The origin of man: A chromosomal pictorial legacy. Science. 215 (4539): 1525-1530.
- Tomkins, J. P. and J. Bergman. 2011. Telomeres: implications for aging and evidence for intelligent design. Journal of Creation. 25 (1): 86-97.
- Ijdo, J. W. et al. 1991. Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proceedings of the National Academy of Sciences. 88 (20): 9051-9055.
- Chaves, R. et al. 2003. Molecular cytogenetic analysis and centromeric satellite organization of a novel 8;11 translocation in sheep: a possible intermediate in biarmed chromosome evolution. Mammalian Genome. 14 (10): 706-710.
- Tsipouri, V. et al. 2008. Comparative sequence analyses reveal sites of ancestral chromosomal fusions in the Indian muntjac genome. Genome Biology. 9 (10): R155.
- Adega, F., H. Guedes-Pinto, and R. Chaves. 2009. Satellite DNA in the karyotype evolution of domestic animals—clinical considerations. Cytogenetic and Genome Research. 126 (1-2): 12-20.
- Bodvarsdottir, S. K. et al. 2012. Dysfunctional telomeres in human BRCA2 mutated breast tumors and cell lines. Mutation Research. 729 (1-2): 90-99.
- Tanaka, H., M. J. Beam, and K. Caruana. 2014. The presence of telomere fusion in sporadic colon cancer independently of disease stage, TP53KRAS mutation status, mean telomere length, and telomerase activity. Neoplasia. 16 (10): 814-823.
- Tu, L. et al. Widespread telomere instability in prostatic lesions. Molecular Carcinogenesis. Published online ahead of print April 27, 2015.
- Fan, Y. et al. 2002. Genomic structure and evolution of the ancestral chromosome fusion site in 2q13-2q14.1 and paralogous regions on other human chromosomes. Genome Research. 12 (11):1651-1662.
- Fan, Y. et al. 2002. Gene content and function of the ancestral chromosome fusion site in human chromosome 2q13-2q14.1 and paralogous regions. Genome Research. 12 (11): 1663-1672.
- Tomkins, J. P. 2013. Alleged Human Chromosome 2 “Fusion Site” Encodes an Active DNA Binding Domain Inside a Complex and Highly Expressed Gene—Negating Fusion. Answers Research Journal. 6: 367-375.
- Tomkins, J. and J. Bergman. 2011. The chromosome 2 fusion model of human evolution—part 2: re-analysis of the genomic data. Journal of Creation. 25 (2): 111-117.
- Azzalin, C. M., S. G. Nergadze, and E. Giulotto. 2001. Human intrachromosomal telomeric-like repeats: sequence organization and mechanisms of origin. Chromosoma. 110 (2): 75-82.
- Simonet, T. et al. 2011. The human TTAGGG repeat factors 1 and 2 bind to a subset of interstitial telomeric sequences and satellite repeats. Cell Research. 21 (7): 1028-1038.
- Yang, D. et al. 2011. Human telomeric proteins occupy selective interstitial sites. Cell Research. 21 (7):1013-1027.
- Heartlein, M. W., J. H. Knoll, and S. A. Latt. 1988. Chromosome instability associated with human alphoid DNA transfected into the Chinese hamster genome. Molecular and Cellular Biology. 8 (9): 3611-3618.
- Archidiacono, N. et al. 1995. Comparative mapping of human alphoid sequences in great apes using fluorescence in situ hybridization. Genomics. 25 (2): 477-484.
- Samonte, R. V., K. H. Ramesh, and R. S. Verma. 1997. Comparative mapping of human alphoid satellite DNA repeat sequences in the great apes. Genetica. 101 (2): 97-104.
* Dr. Tomkins is Research Associate at the Institute for Creation Research and received his Ph.D. in genetics from Clemson University.














