Bacteria sometimes face a rough life. At about a tenth the size of most plant and animal cells, they have no layer of skin to protect them. Environments can change quickly and if microbes don't have the right tools to adapt, they won't last long. Bioengineers modeled three interdependent aspects of a metabolic system that bacteria use to thrive in ever-changing environments, revealing an underlying array of interrelated parts that they described as "underappreciated."1
When biologists seed a fresh batch of sugary broth with C. acetobutylicum bacteria, the first thing those microbes do is harvest the sugar's energy and multiply—their simple method of reproduction. Since no sewage treatment system exists nearby, their organic acid wastes build up around them. But that's no problem for well-equipped bacteria.
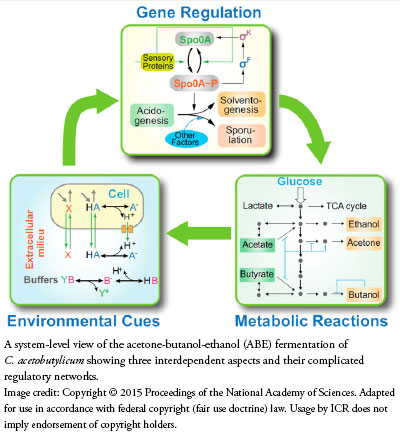
When acids mount up, the bacteria switch on a different internal factory that assimilates those wastes and actually converts them into something useful. Sounds simple from a birds-eye view, but when top scientific minds try to make such a system, they discover that bacterial metabolism is far from simple.
The bacteria use detectors, on/off switches, dimmer switches, relay switches, feedback loops, and a lot of precise engineering to properly connect them—like a complicated computer circuit board. How complicated is it? Just ask the bioengineers who designed a digital version of just one corner of C. acetobutylicum's metabolism. They published their model, which they tested against real bacterial cultures, in Proceedings of the National Academy of Sciences.1
 University of Illinois Engineering News wrote, "Researchers from the University of Illinois at Urbana-Champaign have, for the first time, uncovered the complex interdependence and orchestration of metabolic reactions, gene regulation, and environmental cues of clostridial metabolism."2
University of Illinois Engineering News wrote, "Researchers from the University of Illinois at Urbana-Champaign have, for the first time, uncovered the complex interdependence and orchestration of metabolic reactions, gene regulation, and environmental cues of clostridial metabolism."2
Inside the bacteria, a set of uniquely tailored enzymes (protein machines) facilitates each chemical reaction in a chain of essential metabolic events. Increasing levels of byproducts from this activity makes a toxic and acidic world outside the cell. A second metabolic system saves the bacteria from these potentially lethal byproducts by reconfiguring them.
How does the cell know what its environment contains and what turns on this second metabolic gear? It turns out that tiny machines compare the acidity inside the cell to that outside the cell, and protocols communicate that information to other machines that manage its genes. This is when gene regulation comes into play. This interdependent aspect of the system activates certain genes at just the right time and for just the right durations for the cells to begin converting poisonous organic acids into solvents.
Eventually, the environment becomes too hostile even for these remarkable tactics, at which point the cells switch into preservation mode. Some cells convert themselves into resistant spores that wait until conditions improve before kick starting another growth phase.
At the University of Illinois, bioengineers digitally mimicked subsets of these real-world bacterial systems. They wrote, "The complexity and systems nature of the process have [sic] been largely underappreciated."2 Nothing like reverse-engineering to gain an appreciation for the intricacies built into a real-live system. Reverse-engineering requires intense analysis.
For example, in describing one aspect of building their digital version of the bacteria's gene regulation module, the researchers wrote, "Here, the concentrations of the four key molecules (Spo0A, Spo0A∼P, σF, and σK) were adopted as the main model variables, and their kinetics were described using differential equations."1
Did evolutionary processes really have the ability or forward-thinking insight to construct the metabolism in C. acetobutylicum? Which natural process understood enough basic calculus to integrate differential equations into an essential, dynamic network of interdependent parts and modules? Was it mutations, death of the unfit, or population dynamics?
Since no combination of natural processes fits the bill, an intelligent source must have made bacterial metabolism's interdependent aspects. And that intelligent source must have been even brighter than the excellent engineers who discovered and tried to replicate them. They made a mere digital copy, but He made the real living thing.
References
- Liao, C. et al. Integrated, systems metabolic picture of acetone-butanol-ethanol fermentation by Clostridium acetobutylicum. Proceedings of the National Academy of Sciences. Posted online before print, June 22, 2015, accessed June 29, 2015.
- Kubetz, R. Unlocking fermentation secrets opens the door to new biofuels. Posted on engineering.illinois.edu June 24, 2015, accessed June 25, 2015.
*Mr. Thomas is Science Writer at the Institute for Creation Research.
Article posted on July 20, 2015.



















